utils::available.packages() %>% as_tibble()Diving into dependen-“sea”
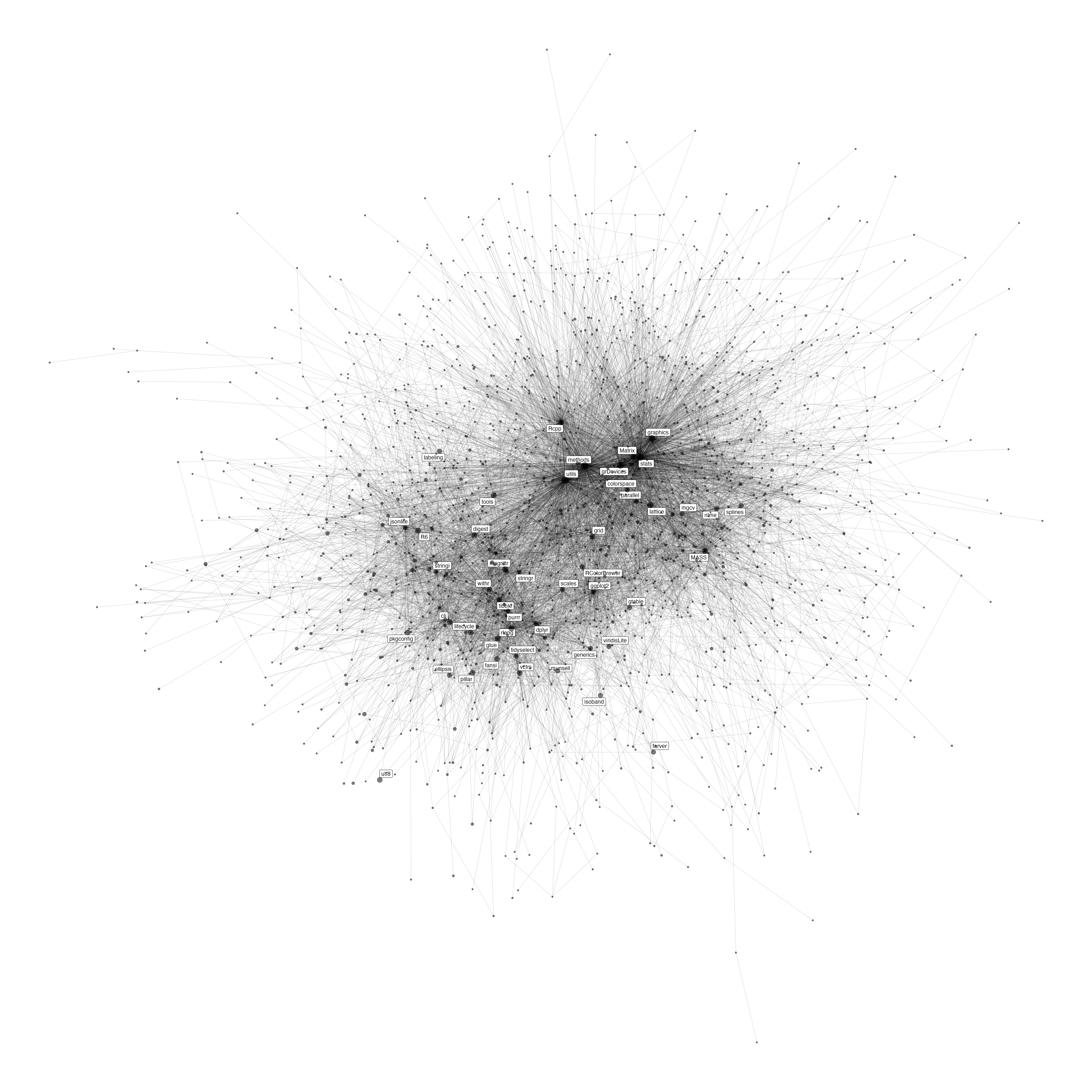
How CRAN packages are interconnected
When writing a package, we may want to use functions in other packages. This creates a dependency for our package and a reverse dependency on the package we borrow functions from. As one of the recipients of the isoband email1, I’m curious to know how interconnected CRAN packages are. Luckily, it is not too hard to get data on this, and so the journey begins…
Preparing dependency data
The utils package provides the function available.packages() to extract CRAN package information. The data includes information on the package name, version, dependency, and license:
code
# A tibble: 18,650 × 17
Package Version Priority Depends Imports LinkingTo Suggests Enhances License
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 A3 1.0.0 <NA> R (>= … <NA> <NA> randomF… <NA> GPL (>…
2 AATtools 0.0.2 <NA> R (>= … magrit… <NA> <NA> <NA> GPL-3
3 ABACUS 1.0.0 <NA> R (>= … ggplot… <NA> rmarkdo… <NA> GPL-3
4 abbrevi… 0.1 <NA> <NA> <NA> <NA> testtha… <NA> GPL-3
5 abbyyR 0.5.5 <NA> R (>= … httr, … <NA> testtha… <NA> MIT + …
6 abc 2.2.1 <NA> R (>= … <NA> <NA> <NA> <NA> GPL (>…
7 abc.data 1.0 <NA> R (>= … <NA> <NA> <NA> <NA> GPL (>…
8 ABC.RAP 0.9.0 <NA> R (>= … graphi… <NA> knitr, … <NA> GPL-3
9 abcADM 1.0 <NA> <NA> Rcpp (… Rcpp, BH <NA> <NA> GPL-3
10 ABCanal… 1.2.1 <NA> R (>= … plotrix <NA> <NA> <NA> GPL-3
# ℹ 18,640 more rows
# ℹ 8 more variables: License_is_FOSS <chr>, License_restricts_use <chr>,
# OS_type <chr>, Archs <chr>, MD5sum <chr>, NeedsCompilation <chr>,
# File <chr>, Repository <chr>From this, we can extract a table to map out the direct dependency every CRAN package has. In this post we will focus on the two strong dependencies: Depends and Imports:
code
all_pkgs <- raw %>%
tidyr::separate_rows(Imports, sep = ",") %>%
tidyr::separate_rows(Depends, sep = ",") %>%
mutate(
across(c(Depends, Imports), ~gsub("\\(.*\\)", "\\1", .x)),
across(c(Depends, Imports), str_trim)
)
(dep_lookup_tbl <- all_pkgs %>%
dplyr::select(Package, Depends, Imports) %>%
rename(downstream = Package) %>%
pivot_longer(Depends:Imports, names_to = "type", values_to = "upstream") %>%
distinct() %>%
filter(!upstream %in% c("R", "")) %>%
filter(!is.na(upstream)) %>%
arrange(downstream))# A tibble: 96,576 × 3
downstream type upstream
<chr> <chr> <chr>
1 A3 Depends xtable
2 A3 Depends pbapply
3 AATtools Imports magrittr
4 AATtools Imports dplyr
5 AATtools Imports doParallel
6 AATtools Imports foreach
7 ABACUS Imports ggplot2
8 ABACUS Imports shiny
9 ABC.RAP Imports graphics
10 ABC.RAP Imports stats
# ℹ 96,566 more rowsDependency is a transitive relation. This means a package also (indirectly) depends on all the dependencies of the package of it imports and so on. Changes from an package will propagate downwards through its dependency chain. With the direct dependency table above, we can iteratively construct the extended dependency tree:
code
find_all_deps <- function(upstream, data){
print(upstream)
dt <- tibble()
dt2 <- data
i <- 1
while(nrow(dt2) > nrow(dt)){
print(i)
dt <- dt2
n <- paste0("upstream", i)
dt2 <- dt %>%
rename(upstream = downstream) %>%
left_join(dep_lookup_tbl %>% select(-type), by = "upstream") %>%
rename(!!quo_name(n) := upstream)
i <- i + 1
}
dep <- dt2 %>%
pivot_longer(
cols = c(contains("upstream"), "downstream"),
names_to = "dump", values_to = "downstream") %>%
distinct(downstream) %>%
filter(!is.na(downstream)) %>%
mutate(downstream = sort(downstream))
return(dep)
}
dep_all <- dep_lookup_tbl %>%
arrange(-desc(upstream)) %>%
nest(direct_deps = -upstream) %>%
mutate(all_deps = map2(upstream, direct_deps, find_all_deps))
(edges <- dep_all %>%
select(-direct_deps) %>%
unnest(all_deps) %>%
filter(!is.na(upstream), !is.na(downstream)))# A tibble: 550,306 × 2
upstream downstream
<chr> <chr>
1 a4Core nlcv
2 abc abctools
3 abc EasyABC
4 abc ecolottery
5 abc nlrx
6 abc paleopop
7 abc poems
8 abc.data abc
9 abc.data abctools
10 abc.data EasyABC
# ℹ 550,296 more rowsThe plot below shows the number of dependencies and reverse dependencies a package has.
code
nodes <- tibble(id = unique(c(edges$upstream, edges$downstream))) %>%
left_join(edges %>% count(upstream, name = "n_revdep"), by = c("id" = "upstream")) %>%
left_join(edges %>% count(downstream, name = "n_dep"), by = c("id" = "downstream")) %>%
filter(!is.na(id)) %>%
mutate(n_revdep = ifelse(is.na(n_revdep), 0, n_revdep),
n_dep = ifelse(is.na(n_dep), 0, n_dep))
################################################################
# deriving color categories
recommended <- raw %>% filter(Priority == "recommended") %>% pull(Package)
base <- c("base", "compiler", "datasets", "grDevices", "graphics", "grid", "methods", "parallel", "splines", "stats", "stats4", "tcltk", "tools", "translations", "utils")
r_lib_gh <- gh("GET /orgs/{username}/repos", username = "r-lib", .limit = 200)
r_lib <- vapply(r_lib_gh, "[[", "", "name")
r_tidyverse_gh <- gh("GET /orgs/{username}/repos", username = "tidyverse", .limit = 40)
tidyverse <- vapply(r_tidyverse_gh, "[[", "", "name")
nodes <- nodes %>%
mutate(category =
case_when(id %in% tidyverse ~ "tidyverse",
id %in% base ~ "base",
id %in% r_lib ~ "r-lib",
id %in% recommended ~ "recommended",
TRUE ~ "zzz"))
################################################################
# to deal with zero mark after sqrt tranform
# https://github.com/tidyverse/ggplot2/issues/980
mysqrt_trans <- function() {
scales::trans_new("mysqrt",
transform = base::sqrt,
inverse = function(x) ifelse(x<0, 0, x^2),
domain = c(0, Inf))
}
p <- nodes %>%
mutate(tooltip = glue::glue("Pkg: {id}, dep: {n_dep}, revdep: {n_revdep}")) %>%
ggplot(aes(x = n_dep, y = n_revdep)) +
geom_point_interactive(aes(tooltip = tooltip)) +
ggrepel::geom_text_repel(
data = nodes %>% filter(n_revdep > 3100),
aes(color= category, label = id), min.segment.length = 0) +
scale_color_brewer(palette = "Set1") +
scale_y_continuous(breaks = c(0, 50, 200, 500, 1000, 2500, 5000, 7500, 10000, 15000), trans = "mysqrt") +
scale_x_continuous(breaks = c(0, 1, 5, 10, 20, 40, 80, 120, 160, 200), trans = "mysqrt") +
theme(panel.grid.minor = element_blank(),
legend.position = "bottom") +
xlab("Number of dependencies") +
ylab("Number of reverse dependencies")girafe(ggobj = p, width_svg = 16, height_svg = 12)The x and y-axes show the number of dependencies and reverse dependencies of a package. Both coordinates are square root transformed to accommodate for the skewness in both measures. Packages with more than 3100 reverse dependencies are labelled. The label color denotes four groups: those in base R, those labelled as “recommended” by CRAN, and those listed in the tidyverse and r-lib organisations on GitHub. Expand the color group below to view the package membership:
color group
| category | packages |
|---|---|
| base | base, compiler, datasets, graphics, grDevices, grid, methods, parallel, splines, stats, stats4, tcltk, tools, utils |
| tidyverse | blob, dbplyr, dplyr, dtplyr, forcats, ggplot2, glue, googledrive, googlesheets4, haven, hms, lubridate, magrittr, modelr, multidplyr, nycflights13, purrr, readr, readxl, reprex, rvest, stringr, tibble, tidyr, tidyverse, vroom |
| r-lib | archive, askpass, available, backports, bench, brio, cachem, callr, carrier, cli, clisymbols, clock, commonmark, conflicted, coro, covr, cpp11, crayon, credentials, debugme, desc, devtools, downlit, ellipsis, err, evaluate, fastmap, filelock, fs, gargle, generics, gert, gh, gitcreds, gmailr, gtable, here, httr, httr2, isoband, jose, keyring, later, lifecycle, lintr, liteq, lobstr, memoise, mockery, pak, pillar, pingr, pkgbuild, pkgcache, pkgconfig, pkgdepends, pkgdown, pkgload, prettycode, prettyunits, processx, progress, ps, R6, ragg, rappdirs, rcmdcheck, rematch2, remotes, rex, rlang, roxygen2, rprojroot, scales, sessioninfo, showimage, slider, sodium, styler, svglite, systemfonts, testthat, textshaping, tidyselect, tzdb, urlchecker, usethis, vctrs, waldo, whoami, withr, xml2, xmlparsedata, xopen, ymlthis, zeallot, zip, roxygen2md, diffviewer, vdiffr, asciicast, cliapp, decor, meltr, sloop, tracer, io, conf, webfakes |
| recommended | boot, class, cluster, codetools, foreign, KernSmooth, lattice, MASS, Matrix, mgcv, nlme, nnet, rpart, spatial, survival |
The plot is interactive so you can hover over points of your interest to read the package name and its numbers of (reverse) dependency.
It’s okay to be a couch Pareto
As you would have already noticed, the distribution of the number of reverse dependencies is highly skewed, even after the square root transformation. To better visualise how the lower number of reverse dependency is distributed, we can plot its cumulative distribution:
code
Code
prct_tbl <- purrr::map_dfr(
unique(nodes$n_revdep) %>% sort(),
~tibble(n = .x, prct = nrow(filter(nodes, n_revdep <= .x)) /nrow(nodes)))
tgt_pnts <- prct_tbl %>%
mutate(p = round(prct, digits = 3)) %>%
filter(p %in% c(0.9, 0.95, 0.99, 0.995, 0.999) | prct == 1 | n %in% c(0, 1, 5)) %>%
group_by(p) %>%
filter(n == min(n))
p2 <- prct_tbl %>%
ggplot() +
geom_line(aes(x = n, y = prct)) +
geom_point(data = tgt_pnts, aes(x = n, y = prct)) +
geom_label(data = tgt_pnts, aes(x = n, y = prct + 0.01, label = p)) +
scale_x_continuous(breaks = round(c(0:3, 10, 100, tgt_pnts$n, max(prct_tbl$n)))) +
coord_trans("pseudo_log") +
theme(panel.grid.minor = element_blank()) +
ylab("Percentage of CRAN pkgs with <= n reverse dependencies")p2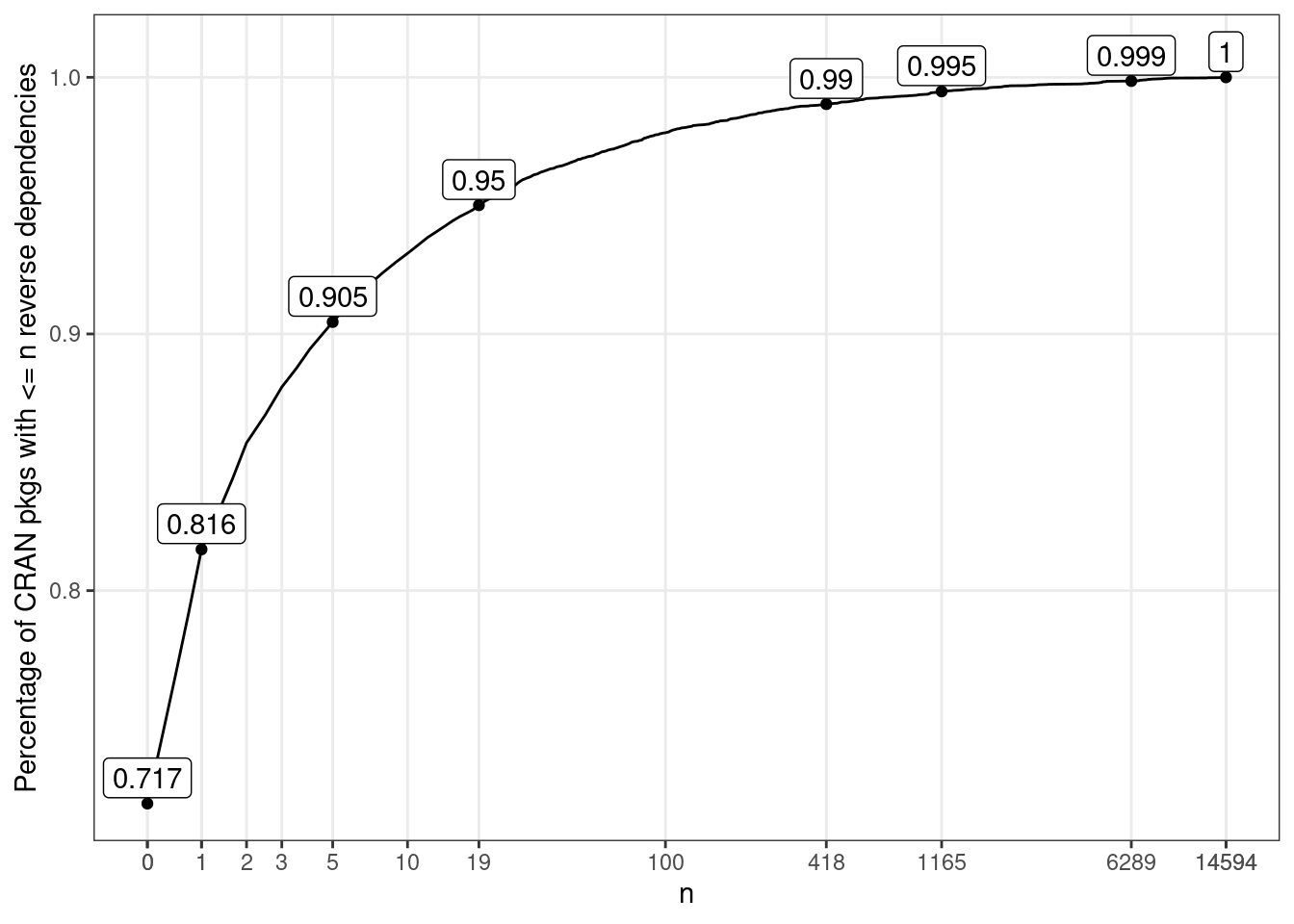
Whether you have guessed or not:
- 73.9% of CRAN packages don’t have any reverse dependency;
- fewer than 10% of the packages on CRAN have more than 5 reverse dependencies; and
- only 1% of the packages have more than 300 reverse dependencies
So while the majority of the R packages do not need reverse dependency checks, a small number of core packages need to test against hundreds or even thousands of reverse dependencies for every new release.
Alternatively, we can rank the packages by their number reverse dependencies (the package with the largest number of reverse dependencies is ranked first). The advantage of this is that there turns out to be a distribution that can capture the shape well: the Zipf–Mandelbrot distribution, the generalised zipf distribution, which is commonly used to model corpus frequency in linguistics:
code
Code
nodes_rank <- nodes %>%
mutate(rank = rank(-n_revdep, ties.method = "first")) %>%
filter(n_revdep > 0)
dt_pos <- nodes %>% filter(n_revdep > 0) %>% pull(n_revdep) %>% sort(decreasing = TRUE)
pred1 <- fitrad(dt_pos, "mand") %>% radpred()
fitted <- tibble(
rank = pred1$rank,
mand = pred1$abund,
count = dt_pos)
p3 <- nodes_rank %>%
ggplot(aes(x = rank, y = n_revdep)) +
geom_point_interactive(aes(tooltip = id)) +
geom_line(data = fitted, aes(x = rank, y = mand), color = "#314f40") +
ggrepel::geom_text_repel(
data = nodes_rank %>% filter(n_revdep > 3100),
aes(label = id, color = category),
min.segment.length = 0) +
scale_y_continuous(breaks = c(10, 50, 200, 500, 1000, 2500, 5000, 7500, 10000, 15000)) +
scale_x_continuous(breaks = c(0, 1, 10, 50, 100, 200, 500, 1000, 2000, 5000, 10000, 20000)) +
coord_trans(x = "sqrt", y = "sqrt") +
scale_color_brewer(palette = "Set1") +
theme_bw() +
theme(panel.grid.minor = element_blank(),
legend.position = "bottom") +
ylab("number of reverse dependencies")girafe(ggobj = p3, width_svg = 16, height_svg = 12)Contender for the next isoband
We can also find packages with similar characteristics as isoband: those with a huge number of reverse dependencies (n > 3000) while not managed by base R or RStudio or listed as recommended on CRAN:
| Package | # of reverse dependency |
|---|---|
| Rcpp | 8376 |
| fansi | 6763 |
| utf8 | 6763 |
| digest | 6394 |
| colorspace | 5028 |
| RColorBrewer | 5017 |
| viridisLite | 4882 |
| farver | 4862 |
| labeling | 4852 |
| munsell | 4851 |
| stringi | 4556 |
| jsonlite | 4295 |
| yaml | 3012 |
| mime | 3005 |
In a quick glance, these packages fall into three categories:
- Color packages:
colorspace,RColorBrewer,viridisLite,munsell - Super giants:
Rcppfor interfacing to C++ for fast computation;jsonlitefor parsing JSON; andstringifor string manipulation.
- Hidden dependencies: These packages don’t have a huge number of direct dependencies themselves but is imported by a giant package:
- imported by
ggplot2:digestfor hashing object in R, andfarverandlabellingare imported byscales, which in turn is imported byggplot2
- imported by
tibble:fansifor ANSI text formatting, andutf8for processing UTF encoding, imported bypillar, which is imported bytibble
- imported by
shiny:mimefor converting file name extension, and - imported by
knitr:yamlfor YAML text conversion
Footnotes
On 5th Oct, CRAN sent out a massive email to inform 4747 downstream package maintainers of the potential archive of package
isobandon 2022-10-19.↩︎